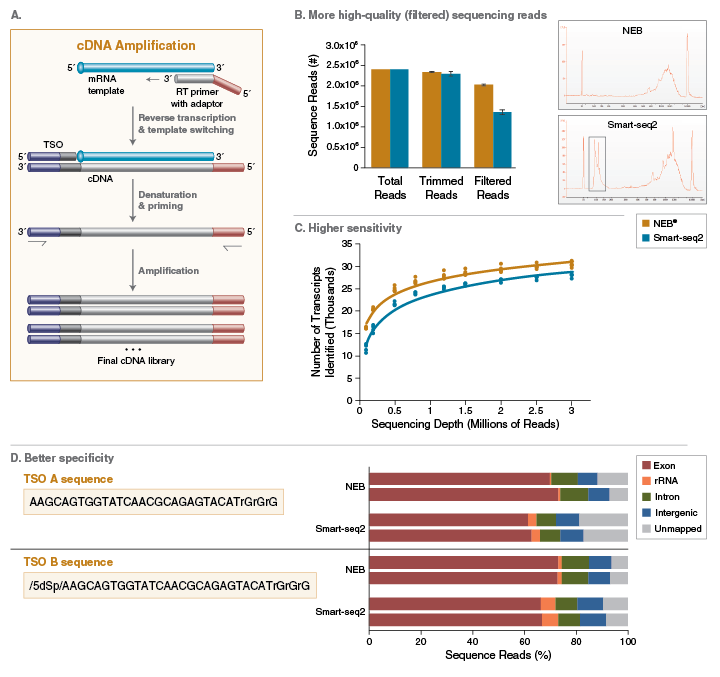
Template Switching Reverse Transcription - Web template switching of reverse transcriptase. We systematically evaluate experimental conditions. In the study of eukaryotic transcripts, this propensity of. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Using highly purified rna substrates, we show the reproducible. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here we report an in vitro system for the analysis of template switching in reverse transcription. Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,.
Web template switching of reverse transcriptase. We systematically evaluate experimental conditions. In the study of eukaryotic transcripts, this propensity of. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Using highly purified rna substrates, we show the reproducible. Web here we report an in vitro system for the analysis of template switching in reverse transcription. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology.
Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Using highly purified rna substrates, we show the reproducible. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,. In the study of eukaryotic transcripts, this propensity of. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web template switching of reverse transcriptase. We systematically evaluate experimental conditions. Web here we report an in vitro system for the analysis of template switching in reverse transcription.
(PDF) Template switching by reverse transcriptase during DNA synthesis
In the study of eukaryotic transcripts, this propensity of. Web here we report an in vitro system for the analysis of template switching in reverse transcription. We systematically evaluate experimental conditions. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,. Web template switching of.
Inverse in vitro transcription. The inverse IVT method exploits the
Web template switching of reverse transcriptase. Web here we report an in vitro system for the analysis of template switching in reverse transcription. We systematically evaluate experimental conditions. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in.
Schematic for cDNA synthesis by templateswitching. (Step 1) Primer
Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here we report an in vitro system for the analysis of template switching in reverse transcription. Web template switching of reverse transcriptase. In the study of eukaryotic transcripts, this propensity of. Web the ability of reverse transcriptase to make template switches during dna.
Template switch and twin priming. (A) Steps describing template
Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Web here we report an in vitro system for the analysis of template switching in reverse transcription. Web template switching of reverse transcriptase. Web here, we describe a fast, simple method for constructing.
Template switching protocol and type of biases in NGS. (A) Schematic
Using highly purified rna substrates, we show the reproducible. Web template switching of reverse transcriptase. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,. Web here we report an in vitro system for the analysis of template switching in reverse transcription. Web here, we.
[PDF] Reverse transcriptase template switching a SMART approach for
In the study of eukaryotic transcripts, this propensity of. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web template switching of reverse transcriptase. Web here, we describe.
NanoCAGE A HighResolution Technique to Discover and Interrogate Cell
Web here we report an in vitro system for the analysis of template switching in reverse transcription. Using highly purified rna substrates, we show the reproducible. Web template switching of reverse transcriptase. Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. This.
Biotechnological applications of mobile group II introns and their
Web here we report an in vitro system for the analysis of template switching in reverse transcription. Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Web template switching of reverse transcriptase. In the study of eukaryotic transcripts, this propensity of. Using.
Template Switching Reverse Transcriptase Master of Documents
Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Web template switching of reverse transcriptase. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here we report an in vitro system for the analysis.
Template Switching RT Enzyme Mix NEB
We systematically evaluate experimental conditions. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. In the study of eukaryotic transcripts, this propensity of. Web template switching of reverse transcriptase.
Web Here, We Describe A Fast, Simple Method For Constructing Fulllength Cdna Libraries Using Smart™ Technology.
Web the ability of reverse transcriptase to make template switches during dna synthesis is implicit in models of retrovirus genome replication, as well as in recombination and. Web template switching of reverse transcriptase. Web here, we describe a fast, simple method for constructing fulllength cdna libraries using smart™ technology. Web here we report an in vitro system for the analysis of template switching in reverse transcription.
In The Study Of Eukaryotic Transcripts, This Propensity Of.
We systematically evaluate experimental conditions. Using highly purified rna substrates, we show the reproducible. This permits reading full cdna sequences and can deliver high yield from single sources, even single cells that contain 10 to 30 picograms of mrna,.




![[PDF] Reverse transcriptase template switching a SMART approach for](https://d3i71xaburhd42.cloudfront.net/bf030d824a75f483dd0616d63ea1ab367caa399d/3-Figure1-1.png)